The HRP’s Annual Research Projects (2016–2021)
In early 2021, the HRP reorganized its consortium members into three working groups. Please select a year or researcher below to see details about specific yearly research projects before this reorganization. We also have an archive of Hearing Health magazine stories about bioinformatics and other HRP updates below.
Comparison of Three Reprogramming Cocktails in the Organ of Corti: Cells, Transcriptomes, and Epigenomes
Andy Groves, Ph.D., Baylor College of Medicine
Each cell type in the human body is defined by its activation of a unique combination of genes that endow each cell type with unique properties. The activation of these genes is achieved by special proteins known as transcription factors, or “switches,” responsible for switching on appropriate genes in one cell type and preventing inappropriate genes from being activated. In recent years, investigators have identified a number of these transcription factors that cause the formation of hair cells. The goal this year is to examine why these cocktails of transcription factors are able to “reprogram” nonsensory cells, but not supporting cells of the inner ear, to become hair cells.
Detection of Transcriptome Changes in Single Cells After Aminoglycoside-Induced Hair Cell Loss in the Chicken Basilar Papilla
Stefan Heller, Ph.D., Stanford University
Birds robustly regenerate their cochlear hair cells through the conversion of dormant supporting cells into new hair cells. Our project uses selective, high-sensitivity methods to reveal the molecular changes in supporting cells after their activation by, for example, ototoxic drugs that cause hair cell death. By examining the responses of many single cells, we have begun to identify triggers that initiate, execute, sustain, and ultimately terminate the regenerative process. Recent experiments have confirmed what we already knew, that the two halves of the chicken cochlea (neural and abneural) predominantly use different regeneration mechanisms. Using bioinformatics methods to process the resulting data, this year we are focusing on analyzing chicken cochlea cell populations isolated at various time points during hair cell regeneration and specifically characterizing the two distinct responses. By examining regeneration in an animal that replaces hair cells after damage, we will be able to find triggers that may be activated in mammals to reverse hair cell loss.
Epigenetics Analysis of Maturation and Regenerative Responses in the Mouse Organ of Corti and Utricle
Neil Segil, Ph.D., University of Southern California
Andy Groves, Ph.D., Baylor College of Medicine
The broad premise of the HRP is to identify molecules that could control hair cell regeneration. To do this, we are studying cell types and regenerative processes in multiple contexts and species, then integrating these data together to identify mechanisms that could potentially be turned on in the mouse cochlea to drive transdifferentiation (activating the correct set of hair cell–promoting genes in supporting cells). The role of this systems biology project is to provide the necessary data integration “glue,” binding together the results from the data generation projects. We will combine much of the data that is being generated by the HRP to advance our knowledge of hair cells, supporting cells, conversion of one cell type to another, and the potential for regeneration. By modeling all of the available HRP data, we will identify regulatory molecules that may contribute to regeneration.
Implementing the gEAR for Data Sharing Within the HRP
Ronna Hertzano, M.D., Ph.D., University of Maryland School of Medicine
One of the successes of the HRP has been the development of the gEAR portal (gene Expression Analysis Resource, umgear.org). The gEAR has many public and private datasets, and these complex datasets can be compared by scientists without the need for sophisticated programming expertise. The gEAR is also the primary data sharing, visualization, and analysis tool for auditory researchers outside of the HRP, becoming a platform that supports the hearing research community at large. This year we will build on past successes, continuing to support data upload, develop new visualization tools, and further enable the greater research community to exploit this resource.
Mouse Model Systems to Interrogate Candidate Genes for Sensory Hair Cell Regeneration
John Brigande, Ph.D., Oregon Health & Science University
As HRP scientists detect and characterize genes that are hypothesized to participate in the activation or inhibition of hair cell regeneration, methods for altering or disrupting those genes are critical to the demonstration of their importance. This project aims to couple together two sophisticated methods for manipulating genes in mice, the so-called CRISPR-READI and i-GONAD methods. CRISPR-READI enables efficient, large gene edits, and i-GONAD simplifies the delivery of reagents for gene editing. Together, the proposed CRISPR-READI-GO method should allow for rapid and efficient gene editing to be put to use by HRP investigators.
Integrative Systems Biology of Hearing Restoration
Seth Ament, Ph.D., University of Maryland School of Medicine
This project will focus on integrating multiple datasets from the HRP to gain insight into hair cell development and regeneration and prioritize specific "driver" genes that can be targeted to induce regeneration. The main premise is that we will be able to regenerate hair cells if we activate the correct set of hair cell–promoting genes in supporting cells. This process is called transdifferentiation, and it occurs naturally in species such as birds and fish, but not in the inner ear of adult mammals. HRP researchers have generated numerous genomic datasets that describe cochlear development and transdifferentiation in multiple species. By analyzing all of these data together using sophisticated network analysis tools, we aim to identify which genes are involved in these processes as well as key differences that may explain the inability of human and mouse cells to transdifferentiate. Finally, this will enable the identification of genes that can be targeted to enable transdifferentiation.
Comparison of Three Reprogramming Cocktails in the Organ of Corti: Cells, Transcriptomes, and Epigenomes
Andy Groves, Ph.D., Baylor College of Medicine
In the past year we have been investigating whether we are able to use genetic reprogramming techniques to generate new hair cells in the mouse cochlea. The results of this work are extremely promising: We were able to turn nonsensory cells of the mouse cochlea into hair cells. However, we consistently find that supporting cells in our mouse models do not respond to reprogramming. This is curious, as supporting cells are the cells responsible for producing new sensory hair cells in birds and fish. In the coming year, we will first examine supporting cells after our reprogramming attempts to see whether they have been able to activate any aspects of a hair cell program. Second, we will test whether hair cell death alters the adjacent supporting cells so that they become more responsive to reprogramming.
Detection of Transcriptome Changes in Single Cells After Aminoglycoside-Induced Hair Cell Loss in the Chicken Basilar Papilla
Stefan Heller, Ph.D., Stanford University
Birds regenerate cochlear hair cells by activating dormant supporting cells. This project builds on innovative methods and findings to study how supporting cells are activated when ototoxic drugs cause hair cell death. The project uses single cell analysis (during which we study the complement of genes that are active in many individual cells) to identify triggers that initiate, execute, sustain, and ultimately terminate the regenerative process. Using bioinformatics methods to process the resulting data, we will focus this year on the analysis of the chicken cochlea cell populations isolated at various time points during hair cell regeneration, which will reveal the molecular steps that occur during hair cell regeneration. We have already identified a first candidate gene signaling pathway that may regulate the regenerative process in the chicken cochlea, and we will be confirming that this pathway plays a role in hair cell regeneration. Interestingly, this pathway is not active during inner ear development, which sets it apart from other pathways linked to chicken (and zebrafish) hair cell regeneration; the other pathways are involved in cell development and may not represent a unique regenerative trigger. Finding triggers that specifically control regeneration may be an important stepping stone on the path to developing cures for hair cell loss in mice and, eventually, humans.
Epigenetics Analysis of Maturation and Regenerative Responses in the Mouse Organ of Corti and Utricle
Neil Segil, Ph.D., University of Southern California
Although hair cell regeneration does not occur in mammals, newborn mice harbor a latent capacity for some regenerative responses. However, this capability disappears within the first few weeks of life. This observation provides an experimental window that this proposal exploits to address fundamental questions about the failure of hair cell regeneration in mammals. Specifically, we propose experiments to identify those changes in the genetic material, the chromatin, that are responsible for orchestrating the differentiation of new hair cells within the newborn organ of Corti in the inner ear, and to investigate the changes in the chromatin that lead to the failure of regeneration in the adult mammalian inner ear.
Implementing the gEAR for Data Sharing Within the HRP
Ronna Hertzano, M.D., Ph.D., University of Maryland School of Medicine
When a group of geographically dispersed scientists collaborate on hair cell regeneration in three different animal models—chicken, zebrafish, and mouse—and use multiple methods to track how genes “instruct” cells (multi-omics), an enormous amount of data results. The work of visualizing, conceptualizing, and analyzing these data presents a considerable challenge, and as technology has advanced, much of the multi-omic data is generated at the single cell level, resulting in datasets and files that are too big to process with traditional tools, such as Excel worksheets. The gEAR portal (gene Expression Analysis Resource, umgear.org) responds to this need by enabling meaningful visualization and analysis of these complex datasets in the public or private domain—no advanced programming skills required. It has also evolved to become a primary data sharing, visualization, and analysis tool for auditory researchers outside of the HRP to become a platform that supports the hearing research community at large.
Mouse Model Systems to Interrogate Candidate Genes for Sensory Hair Cell Regeneration
John Brigande, Ph.D., Oregon Health & Science University
Given what the HRP has discovered thus far, this project proceeds from the assumption that manipulation of multiple genes that regulate diverse signaling pathways may be required to reprogram supporting cells to become functional hair cells. We must first verify the expression pattern of candidate genes in the inner ear, then develop methods that allow us to modulate multiple candidate genes in each supporting cell in order to transmit the genetic instructions that trigger regeneration. This proposal’s first goal is to establish improved genome editing via oviductal nucleic acids delivery (iGONAD) technology, allowing us to identify as well as eliminate candidate genes. Our second goal is to turn on multiple genes simultaneously in the same cell using a mouse model. The overall objective is to establish a rapid method to describe candidate gene expression and function in the inner ear at any stage, and to define an approach to determine which genes are critical to modulate signaling pathways for hair cell regeneration.
Comparison of three reprogramming cocktails
Andy Groves, Ph.D., Baylor College of Medicine
Each cell type in the human body is defined by its activation of a unique combination of genes that endow each cell type with specific properties. The activation of these genes is achieved by special proteins known as transcription factors. These “switches” are responsible for turning on appropriate genes in one cell type and preventing inappropriate genes from being activated. In recent years, investigators have identified a number of these transcription factors that lead to the formation of hair cells in the inner ear. The goal of this project is to rigorously test the extent to which a cocktail of transcription factors is able to reprogram supporting cells of the inner ear to turn into hair cells.
Transcriptome changes in single chick cells
Stefan Heller, Ph.D., Stanford University
Chicks regenerate hair cells in auditory and vestibular organs after damage, making them a valuable animal model to study the signals controlling hair cell regeneration. This project aims to identify changes in gene expression after hair cell loss in the chick cochlea and vestibular system. The collected data will be comprehensive because it will cover all detectable expressed genes. Subsequent data analysis will focus on establishing a sequence of gene expression changes that we hypothesize will correlate with important steps of the hair cell regeneration process. These steps include the signals that initiate, execute, sustain, and ultimately terminate the regenerative process. Comparison among the different organs and across species through collaborations with other HRP investigators will allow us to draw conclusions about species-specific specialized mechanisms as well as more general processes that control hair cell regeneration.
Epigenetics of the mouse inner ear
Michael Lovett, Ph.D., Imperial College London
David Raible, Ph.D., University of Washington
Neil Segil, Ph.D., University of Southern California
Jennifer Stone, Ph.D., University of Washington
Inner ear supporting cells from newborn mice harbor a latent capacity for some regenerative responses, but these disappear within the first few weeks of life. This observation provides an experimental window this proposal will exploit to address fundamental questions about the failure of hair cell regeneration in mammals. Specifically, we propose experiments to identify those changes in the genetic material, the chromatin, that are responsible for orchestrating the differentiation of new hair cells within the perinatal organ of Corti; and investigating the changes in the chromatin, the epigenome, that lead to the failure of regeneration in the adult inner ear.
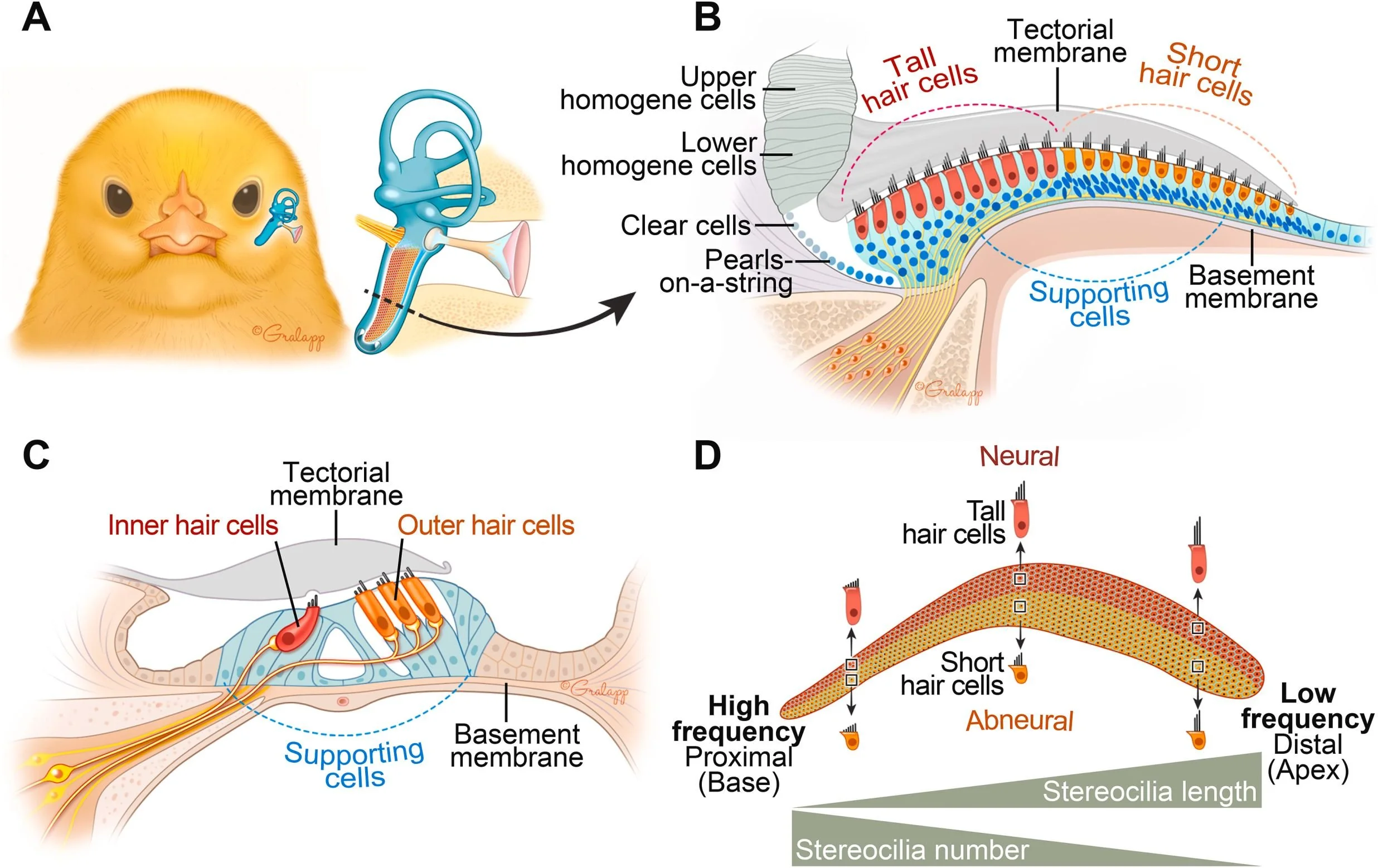
Signaling molecules controlling avian hair cell regeneration
Jennifer Stone, Ph.D., University of Washington
HRP members have spent the past three years gathering information about genes that are turned on or off after inner ear hair cell damage in the chick, fish, and mouse. Some of these genes may encode therapeutic agents that can be applied to stimulate hair cell regeneration in humans. Our HRP studies and others have found that five signaling pathways (Wnt, VEGF, BMP4, Notch, and FGF) are important regulators of hair cell regeneration in the chick cochlea (the basilar papilla). The expression and activity of these pathways change significantly after hair cell damage, and the experimental manipulation of activity in each pathway either boosts or dampens hair cell regeneration. Furthermore, each pathway shows distinct regional expression patterns in the basilar papilla, which implicates it in either mitotic regeneration or non-mitotic regeneration—two distinct ways in which hair cells are replaced after damage. Studies in other growing tissues demonstrate that these five pathways regulate one another in temporally and spatially restricted patterns, in order to coordinate cell growth, differentiation, and patterning. Thus, it is likely that any therapy leading to safe and stable hair cell regeneration will require coordinated manipulation of more than one gene or pathway in the cochlea. In this study, we propose to begin to determine how these five powerful pathways interact to enable and control hair cell regeneration in the chick basilar papilla after hair cell damage.
Integrated systems biology of hearing restoration
Seth Ament, Ph.D., University of Maryland School of Medicine
The goal of this proposal is to support the HRP through data integration and systems biology. We propose two related goals for 2018, based on our preliminary network modeling results and discussions with other HRP investigators. We will (a) predict regulatory genes driving cell fate decisions in the developing mouse cochlea using refined transcriptional regulatory networks, gene co-expression networks, protein-protein interaction networks, and related methods. We will (b) extend these analyses to the zebrafish and chick models by projecting networks from the mouse cochlea onto data from these other species. Our goal is to generate testable predictions about driver genes and perturbations (deviations) that could influence hearing restoration.
Implementing the gEAR for data sharing within the HRP
Ronna Hertzano, M.D., Ph.D. University of Maryland
The HRP takes a multi-investigator, multi-species, multi-omic (methods of tracking gene expression, or instructions), and cell type‐specific approach to define the underpinnings of differences among hair cell regeneration in the chick, fish, and mouse with the aim of identifying keys for hair cell regeneration in mammals. Consequently, the consortium generates large amounts of data that are difficult to visualize, conceptualize, and analyze. The gEAR portal (gene Expression Analysis Resource, umgear.org) allows for simple visualization of multi-omic, multi-species datasets in the public or private domain—without the need for advanced informatics skills. In the first two years of funding from the HRP, we focused primarily on developing tools for multi-omic, multi-species data upload and visualization. Numerous features were added, and all available HRP datasets were uploaded for sharing within the consortium. In parallel, all tools and features developed for the consortium were made available in the public domain—leading the gEAR to be a primary portal for multi-omic data sharing and visualization within the field. With the next two years of funding committed (years three and four), the vast majority of our efforts will be focused on (a) the continued upload of HRP and public datasets, and (b) the development and integration of analysis tools.
Mouse functional testing
John Brigande, Ph.D. Oregon Health & Science University
The conceptual framework of this project wrestles with a persistent challenge facing the HRP consortium: We must verify that the candidate genes we advance as regenerative genes actually perform as advertised. Is our altering of the gene expression of a candidate gene truly the trigger that turns supporting cells into hair cells? Our solution is to devise a mammalian model system that meets several definitive criteria. First, we need deafened adult mammalian inner ears to detect the production of new hair cells; we achieve this genetically by specifically killing hair cells that are uniquely sensitive to a bacterial toxin. Second, we need a way to turn on or turn off the candidate gene after the hair cells are dead; we achieve this by chemically activating a gene that in turn unmasks the expression of the proposed candidate. Third, we need a way to detect newly produced hair cells in the cochlea; we achieve this by using a tissue clearing and staining procedure developed with HRP funding that allows us to detect hair cells produced from supporting cells.
This entire approach is called a model system for validating candidate genes for hair cell regeneration. But one size does not fit all, and we need to continually adapt the core model system to achieve full functionality. In this proposal, we aim to test our model system in healthy ears to see if tweaking our candidate genes can produce hair cells from supporting cells in the absence of widespread hair cell death. The idea here is to make sure that the bacterial toxin–mediated destruction of hair cells is not interfering with our candidate gene activity and new hair cell production. Our second goal is to test a new virus delivery system that will allow us to evaluate larger candidate genes. Presently, we can only express very small genes with the virus we are using, restricting candidate gene verification. Our final goal is to evaluate a modified viral vector that is encased in lipid membranes to learn if it can express candidate genes more efficiently in supporting cells. The benefit of this approach is that viral production is quick, inexpensive, and requires no special training or expertise. Successful completion of this proposal will establish a comprehensive, cost-effective approach to aggressively validate candidate genes for hair cell regeneration.
Identification of candidate regulators of hair cell regeneration in the chick cochlea and utricle
Jennifer Stone, Ph.D. University of Washington
Mark Warchol, Ph.D. Washington University in St. Louis
This project has two components. In the first, the investigators will focus on validation of miRNA-seq data recently acquired by the HRP consortium; these new experiments will determine whether the genomics experiments accurately reflected miRNA fluctuations in the tissue, and may suggest candidate miRNA modulators of hair cell regeneration. In the second component, this project will test which specific signaling pathways are important for the proliferation of supporting cells and the regeneration of hair cells using pharmacological approaches.
Fish epigenomics and enhancer screening
Tatjana Piotrowski, Ph.D. Stowers Institute for Medical Research
As explained in Project #2, many genes are turned off by chemical modifications (epigenetic marks) that silence genes and prevent their activation. This inactivation often occurs at enhancers, which are regions of DNA that control the activation of genes. In this project, Piotrowski will use the ATAC-seq and H3K27ac ChIP-seq methods, which were successful for the mouse inner ear, to find enhancers that are active during hair cell regeneration in the fish. Identification of regeneration enhancers will enable the HRP to examine epigenetic marks comparatively—to determine whether regenerating species, such as the zebrafish and chick, utilize different enhancers than non-regenerating species like the mouse, or whether these enhancers are inactive in mammals.
Fish CRISPR/Cas9 screen, enhancer screen
David Raible, Ph.D. University of Washington
The functional testing of candidate genes is essential for us to be able to wade through the dozens or hundreds of candidates that have been put forward from the Lovett-Warchol RNA-seq experiments. Raible has successfully developed a zebrafish CRISPR screen approach that allows for the rapid testing of candidate genes for their role in hair cell development and regeneration. Raible will continue in his characterization of genes that affect hair cell regeneration. In addition, the project includes a second aim that proposes to test candidate enhancers in the zebrafish.
Integrative systems biology of hearing restoration
Ronna Hertzano, M.D., Ph.D. University of Maryland
The HRP consortium aims to identify factors that can either block or promote regeneration and has generated multiple genomics datasets from inner ears of regenerating and non-regenerating model organisms. This proposal aims to predict these causal factors by a detailed analysis of those datasets. The project’s two investigators—Hertzano, who is well versed in inner ear development and genomics, and Seth Ament, Ph.D., an expert in systems biology and neurobiology—will develop a quantitative model for the gene regulatory networks in hair cells and hair cell precursors that will allow them to predict key genes (e.g., master regulator transcription factors) that are associated with the ability to regenerate hair cells. They will use cutting-edge network biology approaches that integrate information about the preservation and divergence of gene co-expression across species and conditions, with information about the targets of hundreds of transcription factors.
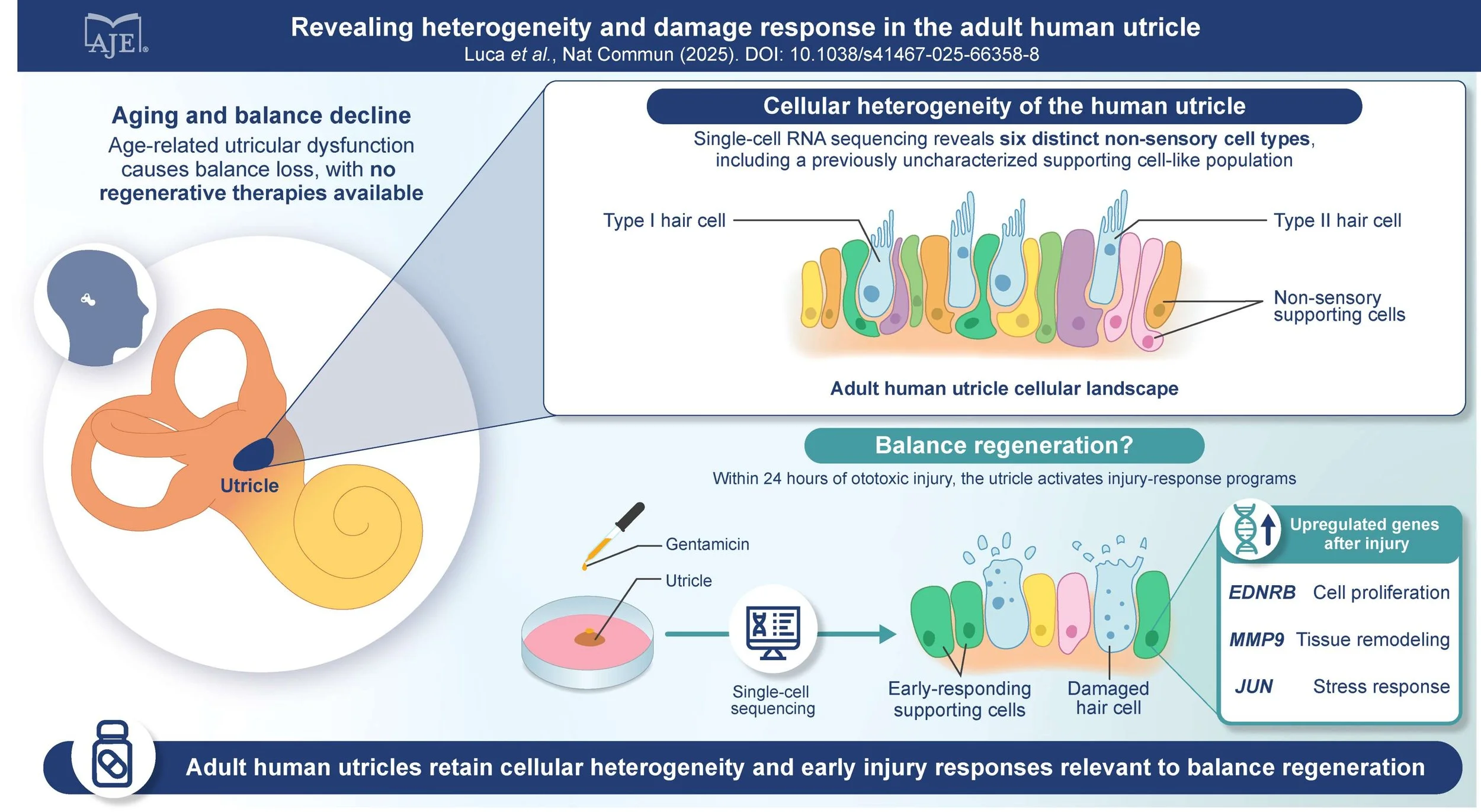
Establishing the human utricle from surgical patients as a translational in vitro model for hair cell regeneration
Alain Dabdoub, Ph.D. University of Toronto
Stefan Heller, Ph.D. Stanford University
Michael Lovett, Ph.D. Imperial College London
These investigators will work with human utricles harvested during surgery to examine whether the response of human inner ear tissue to damage is similar to that of our mammalian model, the mouse. They will culture human utricles for extended lengths of time following damage with aminoglycoside drugs and determine whether the utricles show any proliferation of their supporting cells (making new supporting cells) or regeneration of their hair cells (making new hair cells). Mouse utricles have a limited ability to show proliferation and regeneration, and it is important to determine whether they are a good model for humans. The investigators also intend to examine gene expression in these human utricles to determine the molecular similarity of the cells of interest to their human counterpart cells.
Epigenetics of mouse inner ear maturation and transdifferentiation
Neil Segil, Ph.D. University of Southern California
Jennifer Stone, Ph.D. University of Washington
Michael Lovett, Ph.D. Imperial College London
David Raible, Ph.D. University of Washington
Many genes are turned off by chemical modifications (epigenetic marks) that prevent activation of the gene. One hypothesis is that mammals cannot activate a hair cell regeneration program after the first few postnatal days because the responsible genes have been epigenetically silenced. This second-year project looks at these epigenetic marks in the ear for every gene, during both early and late development. The investigators will use this analysis to find candidate promoter regions, which control gene activity. Preliminary data support the idea that supporting cells turn off expression of key hair cell genes (e.g., Atoh1), and so a plausible approach to triggering regeneration in the mammalian ear is to reverse such changes.
Implementing the gEAR for data sharing within the HRP
Ronna Hertzano, M.D., Ph.D. University of Maryland
Here the gEAR (gene expression for auditory research) portal will be further developed for a second year to perform key gene comparison tasks for the HRP. The gEAR allows for the graphic visualization of gene expression data in an intuitive way: Multiple datasets can be displayed on a single page, all represented by cartoons and dynamically colored based on levels of gene expression. Additional features will be added through dedicated developer time to specifically serve the needs of the consortium, including additional dataset-specific graphics, tools for cross-dataset and cross-species comparisons, and intuitive integration of DNA-structure and gene expression.
Transcriptomics-based analysis of hair cell regeneration in non-mammals
Stefan Heller, Ph.D. Stanford University
Tatjana Piotrowski, Ph.D. Stowers Institute for Medical Research
Jennifer Stone, Ph.D. University of Washington
Mark Warchol, Ph.D. Washington University in St. Louis
Michael Lovett, Ph.D. Imperial College London
In this third-year project, we will disrupt hair cells, then analyze how gene expression changes in single cells from species that show robust hair cell regeneration. Heller’s lab will examine the consequences of aminoglycoside damage at the single-cell level in the chick utricle, while the Piotrowski lab will examine fish lateral-line cells. In another component, the project will add two time points to the chick cochlea and utricle bulk RNA-seq datasets that were generated by Lovett and Warchol and which are extremely valuable datasets for the HRP consortium.
An epigenetic framework for regulating HC regeneration
David Raible, Ph.D. University of Washington
Neil Segil, Ph.D. University of Southern California
Jennifer S. Stone, Ph.D. University of Washington
Many genes are turned off by chemical modifications (epigenetic marks) that prevent activation of the gene. One hypothesis is that mammals cannot activate a hair cell regeneration program after the first few postnatal days because the responsible genes have been epigenetically silenced. This project uses HRP data that looks at these epigenetic marks in the ear for every gene, both during early and late development. The investigators will use this analysis to find candidate promoter regions, which control gene activity. Candidate genetic sequences from the mouse experiments will then be tested in zebrafish. These experiments will allow us to better understand how key hair cell regeneration genes are controlled.
Growing supporting cells in culture: toward high-throughput screens
Alain Dabdoub, Ph.D. University of Toronto
Andy Groves, Ph.D. Baylor College of Medicine
Neil Segil, Ph.D. University of Southern California
There is now a need for experimental systems to quickly test the candidates identified in Phase I experiments before moving into animal models and ultimately clinical trials. These researchers will develop a supporting cell culture system that can be used to quickly screen for factors that promote supporting cell growth or that promote supporting cells to generate hair cells. They will then test if this system can be used to grow cells at low density (allowing fewer cells to be used), to grow cells from older animals (necessary for a real drug), and to grow cells from the adult mouse utricle (which already shows limited regeneration). This project may give us the assay system needed to identify small molecule or gene therapeutics.
Implementing the gEAR for discretionary data sharing within the HRP
Ronna Hertzano, M.D., Ph.D. University of Maryland School of Medicine
Here the gEAR portal (gene Expression for Auditory Research portal) will be adapted to perform key gene comparison tasks for the HRP. The gEAR allows for graphic visualization of gene expression data in an intuitive way: multiple datasets can be displayed in a single page, all represented by cartoons and dynamically colored based on levels of gene expression. Additional features will be added through dedicated developer time to specifically serve the needs of the consortium, including additional dataset-specific graphics, tools for cross-dataset and cross-species comparisons, and intuitive integration of DNA structure and gene expression. Once the gEAR has been adapted for the HRP, all HRP members will be able to browse and interrogate the consortium data on a regular basis, allowing for computational identification of targets that lead to hair cell regeneration.
Bioinformatics
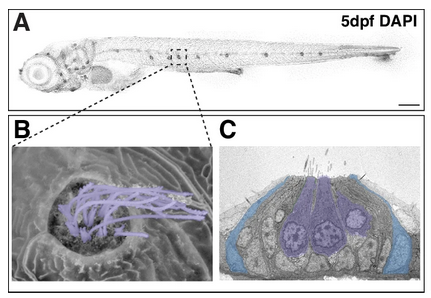
The HRP-funded gEAR (gene Expression Analysis Resource) portal, overseen by HRP scientist Ronna Hertzano, M.D., Ph.D., enables multi-omic data sharing and analysis in the ear field—and recently, has been adapted for neuroscientists as well. This gEAR-created figure shows the development of inner ear sensory cells in the zebrafish embryo five days after fertilization. Its resemblance to North America has made it easier to explain the hair cell regeneration goal of moving south from “Florida” (4) to “Cuba” (2).
Bioinformatics is an emerging discipline that uses information management and analysis tools to uncover relationships between large, disparate sets of data—like what has been generated by the Hearing Restoration Project (HRP). The HRP’s use of bioinformatics not only has uncovered genes that may play roles in hair cell regeneration but also provides an easier way to visualize and understand data.
These stories provide more details:
Sequencing and the HRP, by Stefan Heller, Ph.D., Fall 2013
An Evolving Plan, by Shari Eberts, Spring 2014
Data Mining Toward a Cure, Summer 2014
Transitioning to Phase II, by Peter G. Barr-Gillespie, Ph.D., Fall 2014
Distilling the Data, by Michael Lovett, Ph.D., Summer 2015
Where We Are, and Where We Are Going, by Peter G. Barr-Gillespie, Ph.D., Winter 2016
Hearing Restoration Project Plans Announced for 2016–17, by Peter G. Barr-Gillespie, Ph.D., Summer 2016
The HRP Shifts Gears for Greater Impact, by Peter G. Barr-Gillespie, Ph.D., Winter 2017
Hearing Restoration Project Plans Announced for 2017–18, by Peter G. Barr-Gillespie, Ph.D., Summer 2017
The Hearing Restoration Project Stays the Course, by Peter G. Barr-Gillespie, Ph.D., Winter 2018
Hearing Restoration Project Plans Announced for 2018–19, by Peter G. Barr-Gillespie, Ph.D., Summer 2018
Game Changer, by Ronna Hertzano, M.D., Ph.D., Spring 2018
On a Data-Driven Mission, by Peter G. Barr-Gillespie, Ph.D., Winter 2019
Hearing Restoration Project Plans Announced for 2019–20, by Peter G. Barr-Gillespie, Ph.D., Summer 2019
Data Made Visual, by Christopher Geissler, Ph.D., Spring 2020
Filling in the Gaps, by Peter G. Barr-Gillespie, Ph.D., Spring 2020
The Latest HRP News
We are proud that Hearing Health Foundation-funded scientists are always well represented at Association for Research in Otolaryngology MidWinter Meeting.
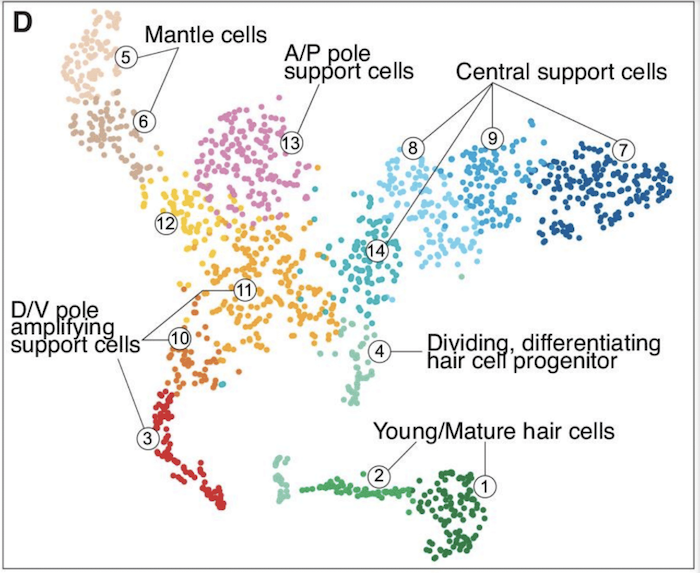
The team’s analysis uncovered a surprising diversity of supporting cells, the “non-sensory cellular guardians” that surround and protect the sensory hair cells and may facilitate their regeneration
Before the HRP, there was no mechanism for data sharing and collaboration, no way to assess gene expression rigorously or to identify relevant patterns, and no examples of new hair cells generated in a post-hearing mammalian cochlea.
The next step in this field is the use of powerful modern sequencing technology in order to map gene activity and gene regulation during hair cell regeneration in fish and birds as well as in mammalian balance organs.
Our mission to fund innovative, groundbreaking hearing and balance science is only possible because of you. We are grateful for the support of our community.
Hearing Restoration Project member and Emerging Research Grants scientist Tatjana Piotrowski, Ph.D., and team show that the transcription factor prdm1a plays a key role in determining hair cell fate in the zebrafish lateral line.
Research has not yet fully explained the mechanisms behind efficient hair cell regeneration in birds, but recent discoveries have sparked multiple promising research directions that might bring us closer to developing treatments for humans.
New research has identified how two distinct genes guide the regeneration of sensory cells in zebrafish. The discovery improves our understanding of how regeneration works in zebrafish and may guide future studies on hearing loss and regenerative medicine in mammals, including humans.
We can now treat otoferlin-related hearing loss. In the next 10 years, we will continue to reach more groups with specific causes of hearing loss—momentum that will help accelerate the process for everyone.
George Arthur Gates, M.D., the inaugural medical director of Hearing Health Foundation’s Hearing Restoration Project (HRP), passed away on February 8, 2025.
As always we are so thrilled to meet and catch up with our funded scientists, past and present, at this important research conference.
This research shows that it is possible to design gene therapies for the ear that are carefully targeted at supporting cells, an essential first step in applying targeted gene therapies to treat hearing loss in humans.











Integrative Systems Biology of Hearing Restoration
Seth Ament, Ph.D., University of Maryland School of Medicine
The broad premise of the HRP is to identify molecules that could control hair cell regeneration. To do this, we are studying cell types and regenerative processes in multiple contexts and species, then integrating these data together to identify mechanisms that could potentially be turned on in the mouse cochlea to drive transdifferentiation (activating the correct set of hair cell–promoting genes in supporting cells). The role of this systems biology project is to provide the necessary data integration “glue,” binding together the results from the data generation projects. We will combine much of the data that is being generated by the HRP to advance our knowledge of hair cells, supporting cells, conversion of one cell type to another, and the potential for regeneration. By modeling all of the available HRP data, we will identify regulatory molecules that may contribute to regeneration.